Minimal Standards Working Group
Purpose
To develop, maintain and promote minimal reporting standards for AIRR-seq data
Members
Co-leaders: Christian Busse and Florian Rubelt
Members: Brian Corrie, Bjoern Peters, Bojan Zimonja, Chaim Schramm, Corey Watson, Encarnita Mariotti-Ferrandiz, Felix Breden, Jean Bürckert, Jerome Jaglale, Lindsay Cowell, Eline Luning Prak, Marie-Paule Lefranc, Nishanth Marthandan, Richard Bruskiewich, Scott Boyd, Scott Christley, Syed Ahmad Chan Bukhari, Uri Hershberg, Steven Kleinstein, Uri Laserson, William Faison
2018 Plans
The WG has identified two lines of work that it will focus on in 2018:
MiAIRR 1.1 (“Make it known, make it easy and demonstrate its utility”)
- Make it known:
- Reach out to and assist other labs to submit their data
- Make it easy:
- Develop toolkit/pipeline for submission and retrieval to/from NCBI
- Evaluate submission to other INDSC repos (EBI/ENA)
- Identify ontologies for a limited number of key data elements (6-8 elements)
- Bug-fix MiAIRR-NCBI implementation
- Refine MiAIRR 1.0 data fields only if necessary, no addition of new data fields
- Demonstrate utility:
- Showcase with a meta-analysis using multiple data sets
MiAIRR 2.0 (“Meeting the needs of the next decade”)
- Develop mechanisms how AIRR-seq studies can report metadata related to clones or singles cells:
- cell phenotypes (e.g. flow cytometry)
- Ig/TCR reactivities and functional properties
- structural information
- Identify strategies to perform zero-knowledge analysis of restricted-access data
Mimimal Standard for Adaptive Immune Receptor Repertoire (MiAIRR)
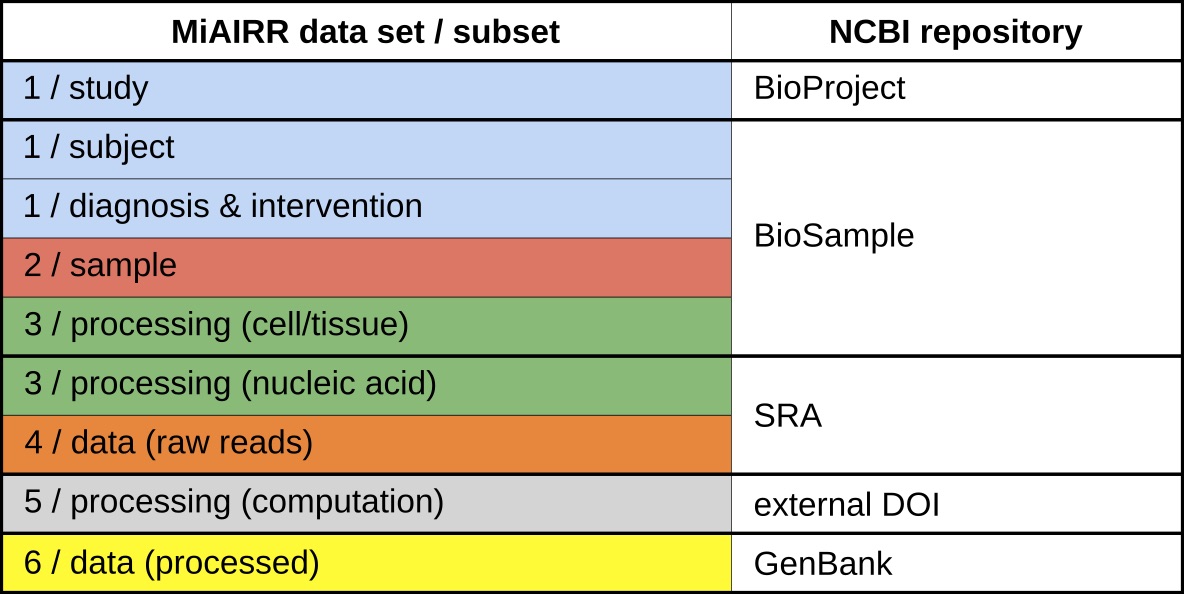
One of the primary initiatives of the AIRR (Adaptive Immune Receptor Repertoire) Community has been to develop a set of metadata standards for the submission of immune receptor repertoire sequencing datasets. In order to support reproducibility, standard quality control, and data deposition in a common repository, the AIRR Community has agreed to six high-level data sets that will guide the publication, curation and sharing of AIRR-Seq data and metadata: Study and subject, sample collection, sample processing and sequencing, raw sequences, processing of sequence data, and processed AIRR sequences. The overall goal of this standard is that sufficient detail be provided such that a person skilled in the art of AIRR sequencing and data analysis will be able to reproduce the experiment and data analyses that were performed.
An implementation of the AIRR data standard has been developed for the NCBI repositories (BioProject, BioSample, Sequence Read Archive (SRA) and GenBank). Each of these repositories stores a subset of the information associated with the six sets as follows:

Do you need help planning the data that you will collect in your experiments? Click here for a detailed description of the data elements and further details
Are you ready to submit your AIRR-seq data? Click here for a detailed “how-to” guide for submission of AIRR-seq data to NCBI databases (BioProject, BioSample, SRA and GenBank).
The development and implementation of the AIRR data standards in NCBI will benefit the whole AIRR community and accelerate widespread use and adoption of immune repertoire research.